Getting started
You can use HippMapp3r through the graphical user interface (GUI) or command line:
For GUI
To start the GUI, type
hippmapper
A GUI that looks like the image below should appear. You can hover any of buttons in the GUI to see a brief description of the command.
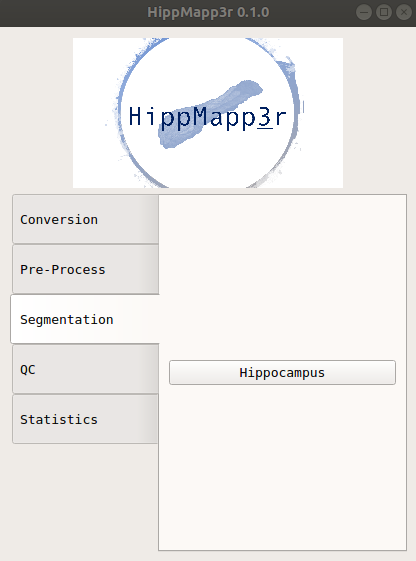
You can get the command usage info by clicking the “Help” box on any of the pop-up windows.
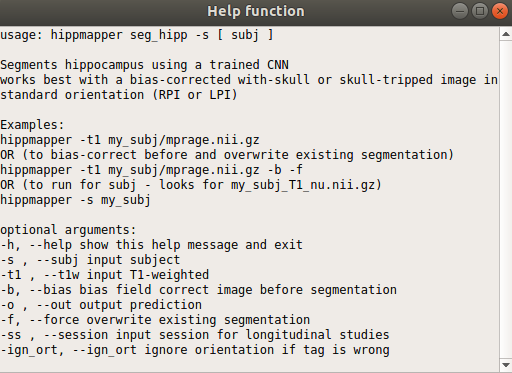
For Command Line
You can see all the hippmapper commands by typing either of the following lines:
hippmapper -h
hippmapper --help
Once you know the command you want to know from the list, you can see more information about the command. For example, to learn more about seg_hfb:
hippmapper seg_hipp -h
hippmapper seg_hipp --help
Hippocampal volumes
To extract hippocampal volumes use the GUI (Stats/Hippocampal Volumes) or command line:
hippmapper stats_hp -h
QC
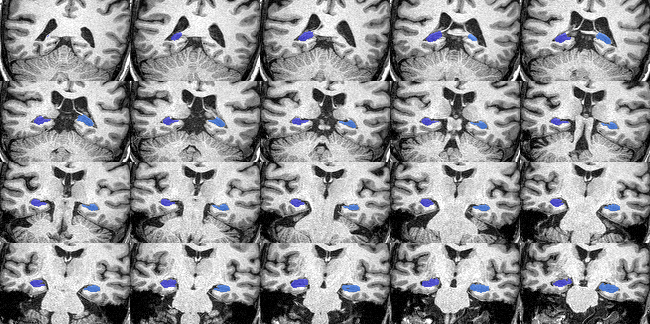
QC files are automatically generated in a sub-folder within the subject folder. They are .png images that show a series of slices in the brain to help you quickly evaluate if your command worked successfully, especially if you have run multiple subjects. They can also be created through the GUI or command line:
hippmapper seg_qc -h
The QC image should look like this:
Logs
Log files are automatically generated in a sub-folder within the subject folder. They are .txt files that contain information regarding the command and can be useful if something did not work successfully.
File conversion
Convert Analyze to Nifti (or vice versa)
hippmapper filetype
Required arguments:
-i , --in_img input image, ex:MM.img
-o , --out_img output image, ex:MM.nii
Example:
hippmapper filetype --in_img subject_T1.img --out_img subject_T1.nii.gz